Troubleshooting ARCHI
TroubleshootingARCHI.RmdIntroduction
ARCHI provides a flexible platform and modeling workflow for regional-scale imputation of hydrologic records. This article highlights some challenges users might face implementing ARCHI and suggests some simple fixes. The following example examines a case where ARCHI fails to impute a dataset.
library(ARCHI)
# Load ARCHI example data
data(LI_data) # Groundwater-level values, in feet below land surface
data(LI_sites) # Site metadata with coordinates referenced to NAD83
# Gridding groundwater levels by monthly medians from 1975 to 2022
grid <- timestep_grid(data = LI_data,
timestep = "monthly",
agg_method = "median",
year_range = c(1975, 2022))
# Trim grid to remove sites with less than 35 percent observed values
grid <- trim_grid(input_grid = grid,
data_thresh = 0.35)
#> 339 site(s) removed with proportion complete less than site-data threshold of 0.35
# Set add_std_means to FALSE (default is TRUE)
mod_fail <- impute_grid(input_grid = grid,
model = "ridge",
error_method = "NSE",
error_thresh = 0,
n_refwl = 10,
sites = LI_sites,
add_std_means = FALSE)
#> Begin initial imputation: 39.46 percent NAs remaining...
#> Iteration 1 complete: 39.46 percent NAs remaining
#> Begin final pass: overwriting initial imputations with improved models...
#> Iteration 2 complete: 39.46 percent NAs remaining
#> Records attempted for imputation = 137
#> Incomplete records remaining = 137
#> Time difference of 0.7256231 secsWhy won’t it work?
There are a few primary reasons why ARCHI may not be able to impute a given dataset. The first is that insufficient correlations exist among the considered records to develop regression models. Another is that the error threshold is too stringent to accept model results. Finally, a common issue is data coverage. ARCHI works best when there are a large proportion of complete or near-complete records present in the initial dataset to help seed the model.
Let’s explore why the above model may have failed. The
mod_notes field of the model_stats data frame
included in the impute_grid output can help us better
understand what happened.
# Create a table showing counts of unique model notes
table(mod_fail$model_stats$mod_notes)
#>
#> Insufficient data in reference records to fill gaps in target record
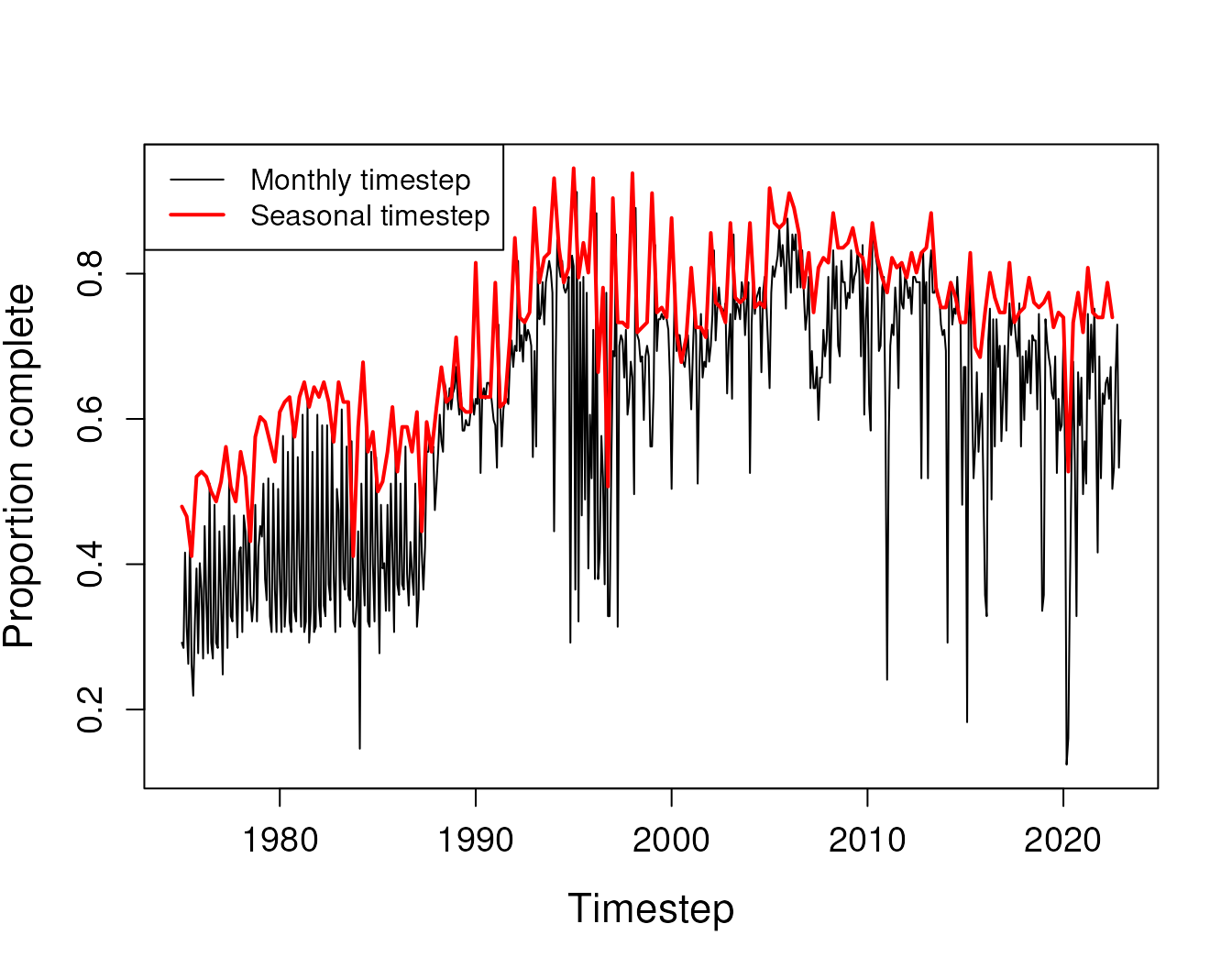
#> 137All 137 sites included in the input grid had insufficient data for gap filling. This is most likely due to periods of the input with very sparse data. Because ARCHI requires references have data to fill all gaps in target records, a handful of data-sparse timesteps may bottleneck expansion of the reference pool. We can see some timesteps have very low proportions of sites with data, particularly March 2020.
# Count number of observations in every row (timestep), excluding the leading timestep index column
prop_tstep <- rowSums(!is.na(grid)[,-1])/(dim(grid)[2]-1)
# The timestep with the least amount of data is March 2020
# Only 12 percent of sites had observed values in that month
prop_tstep[which.min(prop_tstep)]
#> 543
#> 0.1240876
grid$timestep[which.min(prop_tstep)]
#> [1] "2020-03-01"
# Plot the proportion of sites with data for each timestep
plot(grid$timestep, prop_tstep, type = 'l',
xlab = "Timestep", ylab = "Proportion complete",
cex.lab = 1.4, cex.axis = 1.2)
# Add a point at March 2020
points(grid$timestep[which.min(prop_tstep)],
prop_tstep[which.min(prop_tstep)],
cex = 1.5, col = "red", lwd = 2)
# We can also explore data density using the grid_heatmap() function
# plotly::ggplotly creates interactive html widget that allows user to zoom in
# Add a dashed line at March 2020
hm <- grid_heatmap(grid, cb_rng = c(-3,3))
plotly::ggplotly(hm$grid_plot +
ggplot2::geom_vline(xintercept = which.min(prop_tstep),
linetype =3)
)Possible Solutions
In the following sections, we will explore three potential solutions to the data sparsity issue.
1. Add group means
ARCHI can create synthetic references to seed initial imputation
models if the add_std_means input to
impute_grid() is set to TRUE. This is
accomplished by averaging standardized (z-scored by site) observation
values across the dataset or by user-specified groups. By default a
complete reference record named all_sites is generated,
which represents the mean deviation of site records from average
conditions during the period of interest. If a grouping variable is
supplied in the site metadata, each unique group is also used to
generate additional synthetic references, which are attributed with the
user-specified group label. Site groupings selected by the user may be
used to define reference records representative of different geospatial
boundaries or aquifer depth zones.
# Set add_std_means to default of TRUE
mod_out <- impute_grid(input_grid = grid,
model = "ridge",
error_method = "NSE",
error_thresh = 0,
n_refwl = 10,
sites = LI_sites,
add_std_means = TRUE)
#> Begin initial imputation: 39.46 percent NAs remaining...
#> Iteration 1 complete: 0 percent NAs remaining
#> Iteration 2 complete: 0 percent NAs remaining
#> Begin final pass: overwriting initial imputations with improved models...
#> Iteration 3 complete: 0 percent NAs remaining
#> Records attempted for imputation = 137
#> Incomplete records remaining = 0
#> Time difference of 26.4423 secsSuccess! Let’s explore the model output
# Explore model output to find data frame of synthetic references
str(mod_out$std_means)
#> 'data.frame': 576 obs. of 4 variables:
#> $ timestep : Date, format: "1975-01-01" "1975-02-01" ...
#> $ Long_Island: num 0.4531 0.2162 -0.028 -0.0469 -0.1009 ...
#> $ Mainland : num -1.125 -0.952 -1.131 -0.714 -0.492 ...
#> $ all_sites : num 0.019 -0.113 -0.241 -0.217 -0.22 ...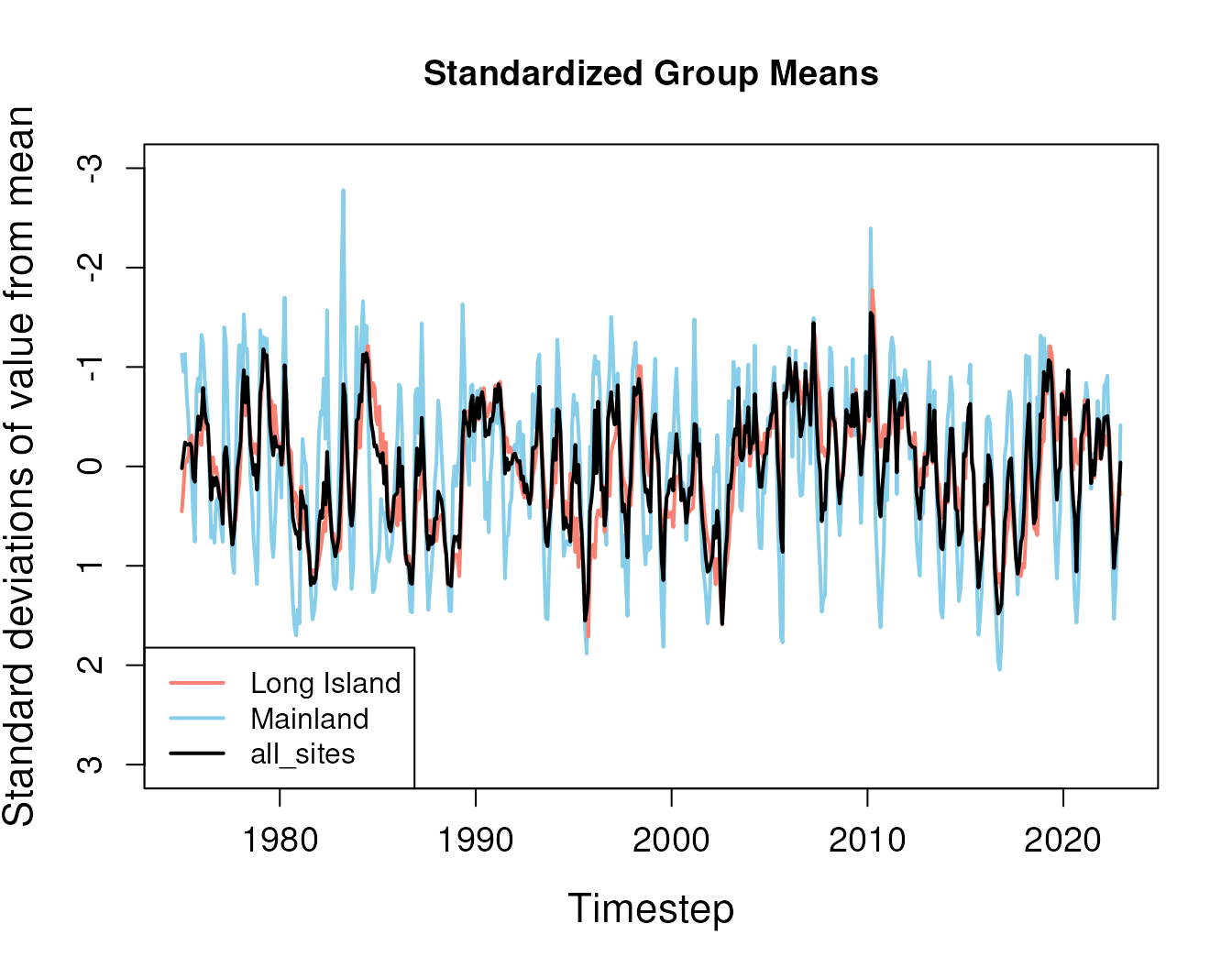 Correlations with these standardized group means can help “seed” the
initial imputation model if few complete records exist.
Correlations with these standardized group means can help “seed” the
initial imputation model if few complete records exist.
2. Truncate period
The modeling period may be adjusted to exclude data-sparse timesteps.
Here, we adjust year_range in the timestep grid function to
exclude 2020.
# Gridding groundwater levels by monthly medians from 1975 to 2019, excluding March 2020
grid_trunc <- timestep_grid(data = LI_data,
timestep = "monthly",
agg_method = "median",
year_range = c(1975, 2019))
# Trim grid to remove sites with less than 35 percent observed values
grid_trunc <- trim_grid(input_grid = grid_trunc,
data_thresh = 0.35)
#> 341 site(s) removed with proportion complete less than site-data threshold of 0.35
# Impute truncated grid
mod_trunc <- impute_grid(input_grid = grid_trunc,
model = "ridge",
error_method = "NSE",
error_thresh = 0,
n_refwl = 10,
add_std_means = F)
#> Begin initial imputation: 38.84 percent NAs remaining...
#> Iteration 1 complete: 0.3 percent NAs remaining
#> Iteration 2 complete: 0 percent NAs remaining
#> Iteration 3 complete: 0 percent NAs remaining
#> Begin final pass: overwriting initial imputations with improved models...
#> Iteration 4 complete: 0 percent NAs remaining
#> Records attempted for imputation = 135
#> Incomplete records remaining = 0
#> Time difference of 23.42125 secsSuccess! We could also use
trim_grid() to remove data sparse timesteps by adjusting
the time_thresh input, but this can create datasets of
irregular frequency that may not accurately reflect time-series dynamics
within a period of interest.
3. Grid coarser timesteps
We can change our timestep frequency from monthly to seasonal (every three months) to improve data coverage.
# Grid groundwater levels at a seasonal timestep from 1975 to 2022
# This aggregates data into a four-season water year beginning in October
grid_seas <- timestep_grid(data = LI_data,
timestep = "seasonal",
agg_method = "median",
type_year = "water",
start_month = 10,
n_seasons = 4,
year_range = c(1975, 2022))
grid_seas$timestep[1:5]
#> [1] "1975-01-01" "1975-04-01" "1975-07-01" "1975-10-01" "1976-01-01"
# Trim grid to remove sites with less than 45 percent observed values
# Approximately same number of sites with sufficient data compared to monthly grid
grid_seas <- trim_grid(input_grid = grid_seas,
data_thresh = 0.45)
#> 330 site(s) removed with proportion complete less than site-data threshold of 0.45We can see that using a seasonal grid for the data has improved
temporal coverage substantially.
Now, let’s impute the seasonal grid.
# Impute seasonal grid
mod_seas <- impute_grid(input_grid = grid_seas,
model = "ridge",
error_method = "NSE",
error_thresh = 0,
n_refwl = 10,
add_std_means = F)
#> Begin initial imputation: 28.3 percent NAs remaining...
#> Iteration 1 complete: 0 percent NAs remaining
#> Iteration 2 complete: 0 percent NAs remaining
#> Begin final pass: overwriting initial imputations with improved models...
#> Iteration 3 complete: 0 percent NAs remaining
#> Records attempted for imputation = 138
#> Incomplete records remaining = 0
#> Time difference of 23.245 secsSuccess!